Introduction to Our Epidemiological Research Activities in the Field
Most of emerging and re-emerging infections as represented by H1N1 Pandemic influenza virus infection, Ebola virus disease, henipavirus infections, Middle East respiratory syndrome, and severe fever with thrombocytopenia syndrome, are zoonoses which cause serious pathology when pathogens carried by natural host animals are transmitted to humans. Therefore, accumulating knowledge of pathogens carried by animals as a preemptive measure against zoonoses which may arise in the future is important. We are involved in epidemiological research on wild animals in the Republic of Zambia and the Republic of Indonesia.
Zambia is located in south-central Africa. It occupies a land area of 752,000 sq km, almost twice the size of Japan, and has a population of 13,470,000, a tenth of that of Japan. In Zambia, there are 73 tribes, and national politics have been stable since the country gained independence. Main industries are agriculture (mainly corn), copper ore mining and tourism. The largest university is the University of Zambia based in Lusaka, the capital city, and has approx. 10,000 students.
The Graduate School of Veterinary Medicine, Hokkaido University and School of Veterinary Medicine, University of Zambia began their relationship in 1985 when the School of Veterinary Medicine was established. This year marks the 30th anniversary of this relationship which became possible through the provision of grant aid from Japan. Along with the Program of Founding Research Centers for Emerging and Re-emerging Infectious Diseases, P2 and P3 laboratories were launched at the School Veterinary Medicine, University of Zambia, and the Hokkaido University Research Center for Zoonosis Control in Zambia was established in August 2008. Since then, various activities have been carried out at this center.
The Division Chief, Professor Sawa has visited the Republic of Zambia more than 40 times to carry out various epidemiological research projects on the following subjects:
Surveillance of Mosquito-Borne Viruses
 Yellow fever virus, dengue virus, Rift Valley fever virus, and chikungunya virus are transmitted to humans and animals through mosquito bites, causing serious infections. To prevent such mosquito-borne infections, vector mosquitoes living in a number of regions in Zambia have been collected, and surveillance of various viruses they carry has been conducted.
Yellow fever virus, dengue virus, Rift Valley fever virus, and chikungunya virus are transmitted to humans and animals through mosquito bites, causing serious infections. To prevent such mosquito-borne infections, vector mosquitoes living in a number of regions in Zambia have been collected, and surveillance of various viruses they carry has been conducted.
Surveillance of Orthopoxviruses
 Monkeypox virus is a species of the genus Orthopoxvirus, and is the causative virus of “monkeypox,” a zoonosis with similar symptoms to that of smallpox and which is prevalent mainly in west and central Africa.
Monkeypox virus is a species of the genus Orthopoxvirus, and is the causative virus of “monkeypox,” a zoonosis with similar symptoms to that of smallpox and which is prevalent mainly in west and central Africa.
We are carrying out surveillance of orthopox viruses in wild animals living in Zambia (Orba Y. et al, J Gen Virol. 2015).
Surveillance of Paramyxoviruses
Paramyxoviruses include a number of viruses, such as measles virus, mumps virus, Nipah virus, RS virus, etc., which cause various diseases in humans.
In recent years, it has been discovered that some of these viruses originated from wild animals.
Hence, we have carried out epidemiological studies on paramyxoviruses in wild animals. Up to now, human parainfluenza virus type 3 (HPIV3), which causes serious respiratory symptoms, infants, young children and elderly people, has been detected from wild nonhuman primate. This virus infection has been shown to be present in wild animals as well (Sasaki M. et al. Emerg Infect Dis, 2013).
Discovery of New Viruses
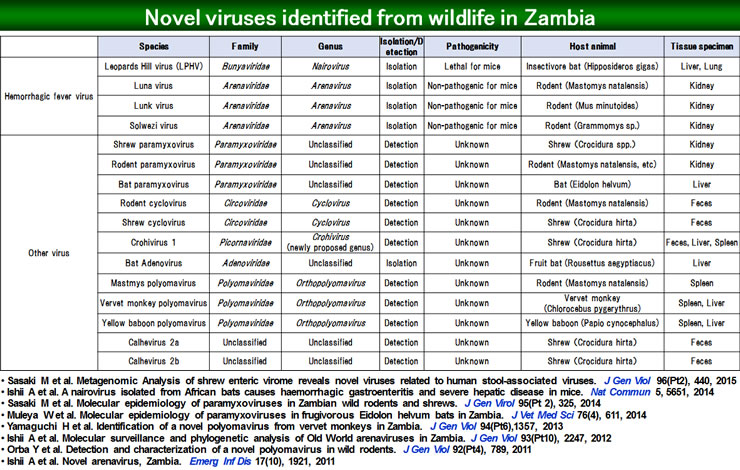
 In the process of seeking out causative viruses of zoonoses, we have discovered a number of new viruses which were previously unknown. The details of these viruses are shown in the figure below (Figure 1).
In the process of seeking out causative viruses of zoonoses, we have discovered a number of new viruses which were previously unknown. The details of these viruses are shown in the figure below (Figure 1).
New types of polyomaviruses were successfully detected from rodent mastomys (Orba Y. et al. J Gen Virol. 2011) and vervet monkeys in Zambia (Yamaguchi H. et al. J Gen Virol. 2013), and from bats captured in Indonesia (Kobayashi S. et al. Arch Virol. 2015).
New types of paramyxoviruses were successfully detected from bats (Walter M. et al. J Gen Virol. 2014) and rodents captured in Zambia (Sasaki M. et al. J Gen Virol. 2014), and from bats captured in Indonesia (Sasaki M. et al. Virol J. 2012).
Genomes of new types of coronaviruses (Anindita PD. et al. Arch Virol. 2015) were detected from bats captured in Indonesia.
Also, new types of herpesviruses were successfully isolated from tissue of bats living in Indonesia, namely fruit bat alphaherpesvirus 1 (FBAHV1). Characteristics of this virus were analyzed. As a result, FBAHV1 was shown to be a herpesvirus closely related to herpesvirus type 1 (HSV-1), and has high pathogenicity in mammals (Sasaki M. et al. J Virol, 2014).
Furthermore, in joint research with Professor Ishii from the Hokkaido University Research Center for Zoonosis Control in Zambia, epidemiological studies on viruses in wild rodent animals (mastomys, African pygmy mouse, etc.) were conducted, and two new viruses, one belonging to a Lassa virus group and is a causative virus of viral hemorrhagic fever, and another belonging to the lymphocytic choriomeningitis virus group, were discovered. These two viruses were named, respectively, Luna virus (Ishii A et al, Emeg Infect Dis, 2011) and Lunk virus (Ishii A et al, J Gen Virol, 2012), after the locations of the host animals.
An exhaustive search was carried out on viruses carried by wild bats which were captured in caves located near to Lusaka, the capital city of Zambia. Wild bats are major natural hosts of pathogens. During analysis, a new virus belonging to the genus Nairovirus in the family Bunyaviridae was discovered and named Leopard Hill virus. Next, an experimental infection study was carried out in mice, in order to analyze the characteristics of the isolated new Nairovirus, Leopard Hill virus. Hepatorenal failure, intestinal tract hemorrhaging, and other symptoms that resemble human hemorrhagic fever caused by the Crimean-Congo hemorrhagic fever virus were successfully reproduced (Ishii A et al. Nat Commun, 2014).
Figure 1. The List of Newly Isolated/Detected Viruses from Wild Animals in Zambia
Viral Metasenomics using Next-Generation Sequencer
Many insectivores, as represented by moles and shrews, live near human-inhabited environments. Their genetic background and feeding behavior differ greatly from those of rodents. While pathogens carried by these animals are expected to also different from those in rodents, little knowledge regarding viruses carried by insectivores is available.
Consequently, the next-generation sequencer was used for a metagenomic analysis on feces of shrews, and an exhaustive search was carried out on viruses present in the feces. As a result, new circovirus and picornavirus, which are closely related to the viruses detected in feces of patients suffering from human acute enteritis and neurological diseases with unknown origin, were discovered. These results suggest that shrews would be potential hosts of pathogens that cause zoonoses (Sasaki et al. J. Gen. Virol., 2015).