Our lab has started from 2005 when the Research Center for Zoonosis Control was established. Many of our students are from overseas, and seminars are conducted in English. The division has both a "wet" environment for molecular biological experiments and a "dry" environment for large-scale genetic information analysis, allowing challenging research using bioinformatics and state-of-the-art equipment.
Our research interest
-We study infectious diseases spread among domestic animals, wild animals, arthropods, and humans.
-We are developing innovative diagnostic methods for pathogens using next-generation sequencers (illumina, Nanopore), and analyzing pathogen evolution and genome function.
-We study protozoa (Toxoplasma, African trypanosomes, Theilelia, Babesia), bacteria (Rickettsia), viruses (metagenomics) and vector arthropods (tsetse flies, ticks, mosquitoes).
Pick up topics
Nanopore sequencer and comprehensive diagnosis of infectious diseases
The next-generation sequencers has revolutionized all life science fields by bringing about an era in which large numbers of nucleotide sequences can be decoded at overwhelmingly low cost. Among these, we are particularly interested in the nanopore sequencers marketed by Oxford Nanopore. This palm-sized, compact sequencer is portable and can be connected to a laptop computer via USB for sequencing. Our goal is to use this low-cost, high-throughput, and portable nanopore sequencer to diagnose new infectious diseases. For example, if pathogen sequences are enriched from patient or animal specimens and comprehensively sequenced, we will be able to understand the infection status of pathogens at a much higher resolution than ever before and determine a more optimal treatment strategy. It is also expected that previously unknown pathogens will be detected. Nanopore sequencers do not require laboratories or expensive equipment, so they can be used anywhere (even in remote African jungles!). We believe that the nanopore sequencer has the potential to become an innovative diagnostic tool for infectious diseases in the future.

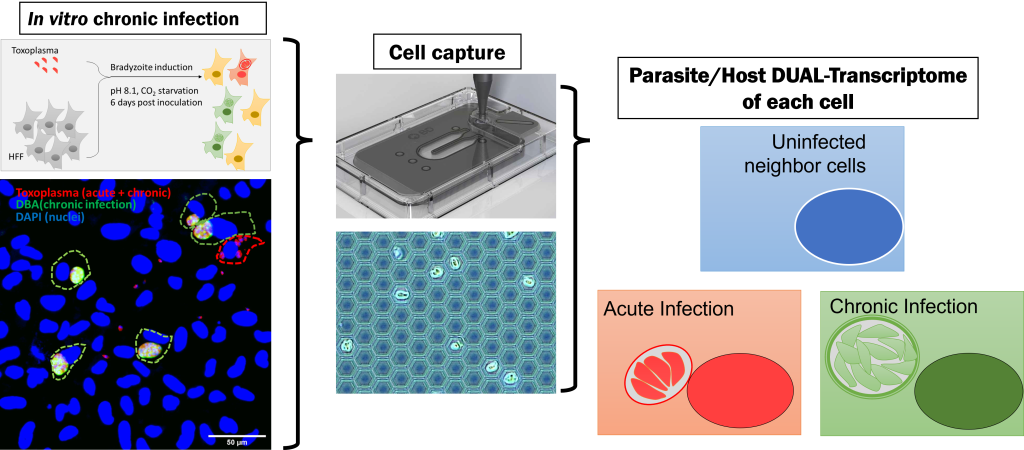
Single cell level transcriptome analysis to understand heterogeneity in host - parasite interaction
Toxoplasma is a zoonotic pathogen that parasitizes mammals, birds, and other animals, including humans, in two distinct phases: an acute infection phase that causes disease and a latent infection phase that may serve as a waiting period for host-to-host transfer and reactivation. It is estimated that approximately 10% to 30% of the world's population is still infected with the disease, although there is some regional variation. The disease is still a problem, especially in immunocompromised patients (HIV-positive AIDS patients) and in pregnancies, where transplacental transmission to the fetus is sometimes fatal. The pathogenesis of Toxoplasma in the acute phase has been actively studied, and many pathogenesis-related factors have been identified, including protozoan factors that act on and regulate the host's immune system. On the other hand, since meat containing cysts in a latently infected state can be a source of infection to humans, it is necessary to know how the latent infection period is established in order to control this disease. It has been reported that the latent infection of Toxoplasma can sometimes last as long as a lifetime, but how the parasite escapes attack by the host's immune system is still unknown. The first step in understanding the response of the immune system to Toxoplasma during the latent infection period is to look at the response of the host cells that are infected with Toxoplasma during the latent infection period. In fact, a sample of cultured cells infected with Toxoplasma is a complex system containing cells infected with Toxoplasma that have reached the latent infection stage, cells containing Toxoplasma in the acute infection stage, and surrounding uninfected cells, making it difficult to observe the "effect of parasite in the latent infection stage". Our laboratory has developed a system in which each cell is divided into small nano-wells and the RNA derived from the cells in the nano-wells is trapped by beads dropped into the nano-wells, making it possible to simultaneously analyze the RNA of the host cell and that of the infecting pathogen at the level of a single infected cell. This analysis has made it possible to simultaneously analyze host cell RNA and RNA of infecting pathogens at the level of a single infected cell. This analysis revealed that the host cellular immune response is activated during the acute infection phase, and that the cellular immune response is also activated in surrounding non-infected cells, but is suppressed in cells with insects in the latent infection phase. We believe that immune system signals that are specifically suppressed during latent infection play a role in evading the host immune system during latent infection, and we are further analyzing the mechanism of this suppression. Not only toxoplasma, but also other parasites interact with host cells at various stages of infection, By applying our analysis at the level of single infected cells, we expect to understand how parasites utilize host cells in each situation and how host cells respond to parasite infection.
On-site diagnostics for Neglected Tropical Diseases
African trypanosomiasis is a zoonosis transmitted to humans from wild and domestic animals via tsetse flies. The disease is one of the so-called Neglected Tropical Diseases (NTDs) because the development of diagnostic and therapeutic agents has been slow due to the fact that the disease is endemic only in Africa. The diagnosis of trypanosomiasis is a matter of concern. Microscopic observation is the standard method for diagnosis of trypanosomiasis, but microscopic observation is difficult to detect when the number of protozoa is small, and observation itself is difficult without a skilled technician. Genetic diagnosis can detect protozoa with high sensitivity and enables detailed species differentiation. To address this issue, our laboratory developed a LAMP diagnostic system in which protozoan genes are amplified and observed as fluorescence when heated at 60-65 degrees Celsius for 30 minutes. Since the reagent is dried using a proprietary method, it can be used even in places where there is no electricity or cold chain (refrigeration/freezing facilities and means of transportation), enabling easy and highly accurate local diagnosis of African trypanosomiasis. This diagnostic technology is being applied to the diagnosis of various tropical diseases.